Submit Raw Reads with Webin-CLI
Introduction
Sequence read data can be submitted to the European Nucleotide Archive (ENA)
using the Webin command line submission interface with -context reads option.
A sequence read submission consists of:
General experiment information
Study accession or unique name (alias)
Sample accession or unique name (alias)
Experiment name
Sequencing platform
Sequencing instrument
Library name (optional)
Library source
Library selection
Library strategy
Free text library description (optional)
Insert size for paired reads (optional)
Read data file(s)
BAM file
CRAM file
Single Fastq file
Paired Fastq files
Multi-fastq files
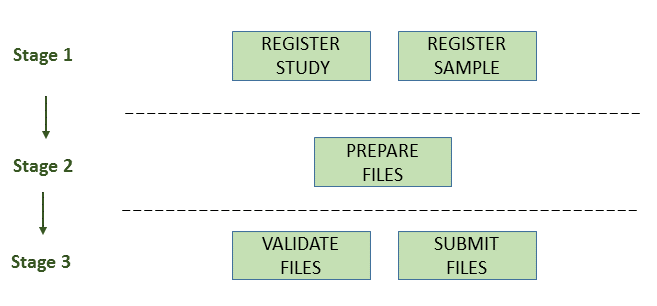
Prepare the files
The set of files that are part of the submission are specified using a manifest file.
The manifest file is specified using the -manifest <filename> option.
A sequence read submission consists of the following files:
1 manifest file
1 BAM file, 1 CRAM file, 1-2 Fastq files or multiple fastq files
Manifest file
The manifest file has two columns and can be submitted in plain text format where the columns are separated by a tab (or any whitespace characters), or in JSON format where the columns are separated by a colon:
Field name (first column): case insensitive field name
Field value (second column): field value
The following metadata fields are supported in the manifest file:
STUDY: Study accession or unique name (alias)
SAMPLE: Sample accession or unique name (alias)
NAME: Unique experiment name
PLATFORM: See permitted values. Not needed if INSTRUMENT is provided.
INSTRUMENT: See permitted values
INSERT_SIZE: Insert size for paired reads
LIBRARY_NAME: Library name (optional)
LIBRARY_SOURCE: See permitted values
LIBRARY_SELECTION: See permitted values
LIBRARY_STRATEGY: See permitted values
DESCRIPTION: free text library description (optional)
Text manifest file format
The following file name fields are supported in the manifest file:
BAM: Single BAM file
CRAM: Single CRAM file
FASTQ: Single fastq file
For example, the following manifest file represents a paired Fastq submission:
STUDY TODO
SAMPLE TODO
NAME TODO
INSTRUMENT Illumina Genome Analyzer II
INSERT_SIZE 200
LIBRARY_SOURCE GENOMIC
LIBRARY_SELECTION RANDOM
LIBRARY_STRATEGY WGS
FASTQ read1.fastq.gz
FASTQ read2.fastq.gz
JSON manifest file format
The JSON manifest file format provides an option to prepare your submission in JSON. This can also be specifically used for more complex data types, such as multi-fastq submissions e.g. for single-cell data.. This is done by entering multiple file names and their respective read_type qualifiers.
The read_type attribute supports the following values:
single
paired
cell_barcode
umi_barcode
feature_barcode
sample_barcode
spatial_barcode
For example, the following manifest file represents a multi-fastq submission:
{
"study": TODO,
"sample": TODO,
"name": TODO,
"platform": "ILLUMINA",
"instrument": "Illumina MiSeq",
"insert_size": "390",
"libraryName": TODO,
"library-source": TODO,
"library_selection": TODO,
"libraryStrategy": TODO,
"fastq": [
{
"value": "single_cell_S1_L001_I1_001.fastq.gz",
"attributes": {
"read_type": "feature_barcode"
}
},
{
"value": "single_cell_S1_L001_R1_001.fastq.gz",
"attributes": {
"read_type": ["paired", "umi_barcode"]
}
},
{
"value": "single_cell_S1_L001_R2_001.fastq.gz",
"attributes": {
"read_type": "sample_barcode"
}
},
{
"value": "single_cell_S1_L001_R3_001.fastq.gz",
"attributes": {
"read_type": ["paired", "cell_barcode"]
}
}
]
}
Metadata validation
Permitted values for platform
BGISEQ: Sequencers based on DNBSEQ by MGI Tech.
CAPILLARY: Sequencers based on capillary electrophoresis technology manufactured by LifeTech (formerly Applied BioSciences).
DNBSEQ: Uses DNA nanoballs(DNB) and regular array chips.
ELEMENT: Element Biosciences uses avidity sequencing: a polymerase inserts nucleotides, detected using fluorescence.
HELICOS: Helicos is similar to 454 technology - uses 1-color sequential flows.
ILLUMINA: 54 technology use 1-color sequential flows: 454 technology use 1-color sequential flows.
ION_TORRENT: Ion Torrent Personal Genome Machine (PGM) from Life Technologies. Directly translates chemically encoded information (A, C, G, T) into digital on semiconductor chip.
LS454: 454 technology use 1-color sequential flows.
OXFORD_NANOPORE: Oxford Nanopore platform type. nanopore-based electronic single molecule analysis.
PACBIO_SMRT: PacificBiosciences platform type for the single molecule real time (SMRT) technology.
ULTIMA: Ultima Genomics platform type. Flowing one nucleotide at a time in order, iteratively.
Deprecated:
ABI_SOLID: ABI is 4-channel flowgram with 1-to-1 mapping between basecalls and flows.
COMPLETE_GENOMICS: CompleteGenomics platform type. At present there is no instrument model. Single-tube long fragment read (stLFR) technology.
Permitted values for instrument
454 GS
454 GS 20
454 GS FLX
454 GS FLX Titanium
454 GS FLX+
454 GS Junior
AB 310 Genetic Analyzer
AB 3130 Genetic Analyzer
AB 3130xL Genetic Analyzer
AB 3500 Genetic Analyzer
AB 3500xL Genetic Analyzer
AB 3730 Genetic Analyzer
AB 3730xL Genetic Analyzer
AB 5500 Genetic Analyzer
AB 5500xl Genetic Analyzer
AB 5500xl-W Genetic Analysis System
BGISEQ-50
BGISEQ-500
DNBSEQ-G400
DNBSEQ-G400 FAST
DNBSEQ-G50
DNBSEQ-T7
Element AVITI
GridION
Helicos HeliScope
HiSeq X Five
HiSeq X Ten
Illumina Genome Analyzer
Illumina Genome Analyzer II
Illumina Genome Analyzer IIx
Illumina HiScanSQ
Illumina HiSeq 1000
Illumina HiSeq 1500
Illumina HiSeq 2000
Illumina HiSeq 2500
Illumina HiSeq 3000
Illumina HiSeq 4000
Illumina HiSeq X
Illumina MiSeq
Illumina MiniSeq
Illumina NovaSeq 6000
Illumina NovaSeq X
Illumina iSeq 100
Ion GeneStudio S5
Ion GeneStudio S5 Plus
Ion GeneStudio S5 Prime
Ion Torrent Genexus
Ion Torrent PGM
Ion Torrent Proton
Ion Torrent S5
Ion Torrent S5 XL
MGISEQ-2000RS
MinION
NextSeq 1000
NextSeq 2000
NextSeq 500
NextSeq 550
PacBio RS
PacBio RS II
PromethION
Sequel
Sequel II
Sequel IIe
UG 100
unspecified
Deprecated:
Complete Genomics
AB SOLiD 3 Plus System
AB SOLiD 4 System
AB SOLiD 4hq System
AB SOLiD PI System
AB SOLiD System
AB SOLiD System 2.0
AB SOLiD System 3.0
Permitted values for library selection
RANDOM: No Selection or Random selection
PCR: target enrichment via PCR
RANDOM PCR: Source material was selected by randomly generated primers.
RT-PCR: target enrichment via
HMPR: Hypo-methylated partial restriction digest
MF: Methyl Filtrated
repeat fractionation: Selection for less repetitive (and more gene rich) sequence through Cot filtration (CF) or other fractionation techniques based on DNA kinetics.
size fractionation: Physical selection of size appropriate targets.
MSLL: Methylation Spanning Linking Library
cDNA: PolyA selection or enrichment for messenger RNA (mRNA); synonymize with PolyA
cDNA_randomPriming:
cDNA_oligo_dT:
PolyA: PolyA selection or enrichment for messenger RNA (mRNA); should replace cDNA enumeration.
Oligo-dT: enrichment of messenger RNA (mRNA) by hybridization to Oligo-dT.
Inverse rRNA: depletion of ribosomal RNA by oligo hybridization.
Inverse rRNA selection: depletion of ribosomal RNA by inverse oligo hybridization.
ChIP: Chromatin immunoprecipitation
ChIP-Seq: Chromatin immunoPrecipitation, reveals binding sites of specific proteins, typically transcription factors (TFs) using antibodies to extract DNA fragments bound to the target protein.
MNase: Identifies well-positioned nucleosomes. uses Micrococcal Nuclease (MNase) is an endo-exonuclease that processively digests DNA until an obstruction, such as a nucleosome, is reached.
DNase: DNase I endonuclease digestion and size selection reveals regions of chromatin where the DNA is highly sensitive to DNase I.
Hybrid Selection: Selection by hybridization in array or solution.
Reduced Representation: Reproducible genomic subsets, often generated by restriction fragment size selection, containing a manageable number of loci to facilitate re-sampling.
Restriction Digest: DNA fractionation using restriction enzymes.
5-methylcytidine antibody: Selection of methylated DNA fragments using an antibody raised against 5-methylcytosine or 5-methylcytidine (m5C).
MBD2 protein methyl-CpG binding domain: Enrichment by methyl-CpG binding domain.
CAGE: Cap-analysis gene expression.
RACE: Rapid Amplification of cDNA Ends.
MDA: Multiple Displacement Amplification, a non-PCR based DNA amplification technique that amplifies a minute quantifies of DNA to levels suitable for genomic analysis.
padlock probes capture method: Targeted sequence capture protocol covering an arbitrary set of nonrepetitive genomics targets. An example is capture bisulfite sequencing using padlock probes (BSPP).
other: Other library enrichment, screening, or selection process.
unspecified: Library enrichment, screening, or selection is not specified.
Permitted values for library source
GENOMIC: Genomic DNA (includes PCR products from genomic DNA).
GENOMIC SINGLE CELL:
TRANSCRIPTOMIC: Transcription products or non genomic DNA (EST, cDNA, RT-PCR, screened libraries).
TRANSCRIPTOMIC SINGLE CELL:
METAGENOMIC: Mixed material from metagenome.
METATRANSCRIPTOMIC: Transcription products from community targets
SYNTHETIC: Synthetic DNA.
VIRAL RNA: Viral RNA.
OTHER: Other, unspecified, or unknown library source material.
Permitted values for library strategy
WGS: Whole Genome Sequencing - random sequencing of the whole genome (see pubmed 10731132 for details)
WGA: Whole Genome Amplification followed by random sequencing. (see pubmed 1631067,8962113 for details)
WXS: Random sequencing of exonic regions selected from the genome. (see pubmed 20111037 for details)
RNA-Seq: Random sequencing of whole transcriptome, also known as Whole Transcriptome Shotgun Sequencing, or WTSS). (see pubmed 18611170 for details)
ssRNA-seq: Strand-specific RNA sequencing.
miRNA-Seq: Micro RNA sequencing strategy designed to capture post-transcriptional RNA elements and include non-coding functional elements. (see pubmed 21787409 for details)
ncRNA-Seq: Capture of other non-coding RNA types, including post-translation modification types such as snRNA (small nuclear RNA) or snoRNA (small nucleolar RNA), or expression regulation types such as siRNA (small interfering RNA) or piRNA/piwi/RNA (piwi-interacting RNA).
FL-cDNA: Full-length sequencing of cDNA templates
EST: Single pass sequencing of cDNA templates
Hi-C: Chromosome Conformation Capture technique where a biotin-labeled nucleotide is incorporated at the ligation junction, enabling selective purification of chimeric DNA ligation junctions followed by deep sequencing.
ATAC-seq: Assay for Transposase-Accessible Chromatin (ATAC) strategy is used to study genome-wide chromatin accessibility. alternative method to DNase-seq that uses an engineered Tn5 transposase to cleave DNA and to integrate primer DNA sequences into the cleaved genomic DNA.
WCS: Random sequencing of a whole chromosome or other replicon isolated from a genome.
RAD-Seq:
CLONE: Genomic clone based (hierarchical) sequencing.
POOLCLONE: Shotgun of pooled clones (usually BACs and Fosmids).
AMPLICON: Sequencing of overlapping or distinct PCR or RT-PCR products. For example, metagenomic community profiling using SSU rRNA.
CLONEEND: Clone end (5’, 3’, or both) sequencing.
FINISHING: Sequencing intended to finish (close) gaps in existing coverage.
ChIP-Seq: ChIP-seq, Chromatin ImmunoPrecipitation, reveals binding sites of specific proteins, typically transcription factors (TFs) using antibodies to extract DNA fragments bound to the target protein.
MNase-Seq: Identifies well-positioned nucleosomes. uses Micrococcal Nuclease (MNase) is an endo-exonuclease that processively digests DNA until an obstruction, such as a nucleosome, is reached.
Ribo-Seq: Ribosome profiling (also named ribosome footprinting) uses specialized messenger RNA (mRNA) sequencing to determine which mRNAs are being actively translated and produces a “global snapshot” of all the ribosomes active in a cell at a particular moment, known as a translatome.
DNase-Hypersensitivity: Sequencing of hypersensitive sites, or segments of open chromatin that are more readily cleaved by DNaseI.
Bisulfite-Seq: MethylC-seq. Sequencing following treatment of DNA with bisulfite to convert cytosine residues to uracil depending on methylation status.
CTS: Concatenated Tag Sequencing
MRE-Seq: Methylation-Sensitive Restriction Enzyme Sequencing.
MeDIP-Seq: Methylated DNA Immunoprecipitation Sequencing.
MBD-Seq: Methyl CpG Binding Domain Sequencing.
Tn-Seq: Quantitatively determine fitness of bacterial genes based on how many times a purposely seeded transposon gets inserted into each gene of a colony after some time.
VALIDATION: CGHub special request: Independent experiment to re-evaluate putative variants.
FAIRE-seq: Formaldehyde Assisted Isolation of Regulatory Elements. Reveals regions of open chromatin.
SELEX: Systematic Evolution of Ligands by Exponential enrichment
RIP-Seq: Direct sequencing of RNA immunoprecipitates (includes CLIP-Seq, HITS-CLIP and PAR-CLIP).
ChIA-PET: Direct sequencing of proximity-ligated chromatin immunoprecipitates.
Synthetic-Long-Read: binning and barcoding of large DNA fragments to facilitate assembly of the fragment
Targeted-Capture: Enrichment of a targeted subset of loci.
Tethered Chromatin Conformation Capture:
OTHER: Library strategy not listed.
CRAM file validation
Reference sequence validation
Reference sequences in CRAM files are required to exists in ENA’s CRAM reference registry.
The Webin command line submission interface maintains two file based caches to avoid unnecessary calls to the registry:
Cache for reference sequence checksums
Cache for reference sequences
The cache for reference sequence checksums is stored in the $HOME/.webin-cli/cram-ref-info directory,
where $HOME is the home directory for the user executing the program.
The cache for reference sequences is configured using the REF_PATH and REF_CACHE environmental
variables as in samtools.
Webin-CLI Validation
When submitting data using the Webin command line interface, your files will be validated and uploaded to your private Webin file upload area in webin.ebi.ac.uk. Any validation error reports are written into the <outputDir>/<context>/<name>/validate directory. Read more about validation reports on the Webin-CLI Submissions section.