Webin-CLI Submission
Introduction
Submissions to ENA can be made using the interactive Webin submission service, programmatic Webin submission service and the Webin command line submission service.
This module gives an introduction to the Webin command line submission interface used to validate, upload and submit files to the European Nucleotide Archive (ENA) and will also link to where you can download it. Please note that unlike with other ENA submissions routes you may have used, you do not need to pre-upload your files when using Webin-CLI.
Webin-CLI is the only way to submit assembled genomes and transcriptomes.
Webin-CLI is available as a Docker image and as a Java jar file.
Download the program as a Java jar file
You can download Webin-CLI Java jar file from its GitHub repository. We recommend always using the latest version:
To get started with running Webin-CLI, download the .jar file for whatever version you’re interested in.
If you have a GitHub account, you can use the ‘Watch’ button in the top right to always be notified of new releases.
Please note that Webin-CLI requires that you have Java installed before you can run it. You should have version 1.8 or newer installed, which can be downloaded from Java:
Webin-CLI has been tested against openjdk version 1.8.0_212. You are recommended to use equivalent or later version.
Download openapi JDKs from the below links:
https://adoptopenjdk.net/?variant=openjdk8&jvmVariant=hotspot
Download Oracle JREs from the below links:
Run the program as a Java jar file
The Webin command line submission interface is a self-executing Java jar file and is
run using the java command:
java -jar webin-cli-<version>.jar <options>
for example:
java -jar webin-cli-1.7.3.jar <options>
The <version> is the version number of the program.
Please note that the command must include the location of the jar file. For example, if you have it in your Downloads directory, the appropriate command on Mac/Linux on immediately opening the terminal would be:
java -jar Downloads/webin-cli-1.7.3.jar <options>
On Windows a backward slash is used instead of a forward slash:
java -jar Downloads\webin-cli-1.7.3.jar <options>
The command line <options> are explained below.
Video Guide: Getting Started With Webin-CLI in Windows 10
Command Line Options
-context: the submission type:-context genome-context transcriptome-context sequence-context reads
-userName: the Webin submission account name.-password: the Webin submission account password.-centerName: the center name of the submitter (mandatory for broker accounts).-manifest: the manifest file name.-outputDir: directory for output files.-inputDir: input directory for files declared in manifest file.-validate: validates the files defined in the manifest file.-submit: validates and submits the files defined in the manifest file.-test: use Webin test service instead of the production service. Please note that the Webin upload area is shared between test and production services, and that test submission files will not be archived.-ascp: use Aspera Cli instead of FTP file transfer, if available. Aspera Cli should be installed and path to executable “ascp” should be in PATH variable.-version: prints the version number of the program and exists.-help: detailed information about the different options.
Submission Process
Please note that this section serves as a general overview of the use of Webin-CLI. You may prefer to find the page specific to your submission type using the links in the sidebar of this page. The following types of submissions are supported:
genome assemblies
transcriptome assemblies
annotated sequences
reads
The type of the submission is specified using the -context command line option:
-context genome-context transcriptome-context sequence-context reads
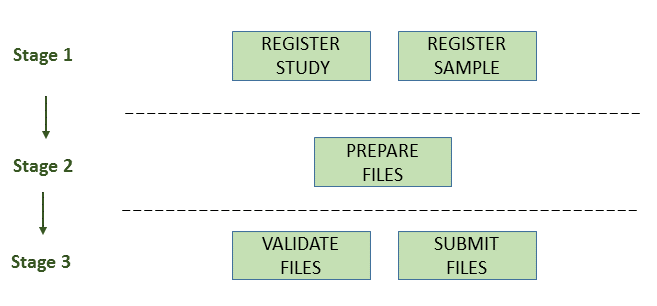
The following picture illustrates the stages of the submission process:
Bulk submissions for reads, unannotated genome assemblies, targeted sequences and taxonomic reference data can be made with Webin-CLI using the following tool:
Webin-CLI Bulk Submissions Tool
Stage 1: Pre-register Study and Sample
Each submission must be associated with a pre-registered study and a sample.
Stage 2: Prepare the Files
The set of files that are part of the submission must be specified using a manifest file.
The manifest file is specified using the -manifest <filename> option.
Manifest File Format
The manifest file can be submitted as either a plain text file or a JSON file.
The manifest file contains metadata fields and file name fields.
Text Manifest File
The text manifest file format has two columns separated by a tab (or any whitespace characters):
Field name (first column): case insensitive field name
Field value (second column): field value
Examples of metadata fields are study and sample references:
STUDY Study accession or unique name (alias)
SAMPLE Sample accession or unique name (alias)
ANALYSIS_REF Comma separated list of analysis accession(s)
RUN_REF Comma separated list of run accession(s)
The file name field format is:
<file type> <file name>
An example of a file name field is:
FASTA genome.fasta.gz
For example, the following manifest file represents a genome assembly consisting of contigs provided in one fasta file:
STUDY TODO
SAMPLE TODO
ASSEMBLYNAME TODO
COVERAGE TODO
PROGRAM TODO
PLATFORM TODO
MINGAPLENGTH TODO
MOLECULETYPE genomic DNA
FASTA genome.fasta.gz
JSON Manifest File
The JSON manifest file format provides an option to prepare your submission in JSON. This can also be specifically used for more complex data types, such as multi-fastq submissions e.g. for single-cell data.
The manifest file has two columns separated by a colon:
Field name (first column): case insensitive field name
Field value (second column): field value
For example, the following manifest file represents a multi-fastq submission for single-cell data:
{
"study": "TODO",
"sample": "TODO",
"name": "TODO",
"platform": "TODO",
"instrument": "TODO",
"insert_size": "TODO",
"library_name": "TODO",
"library_source": "TODO",
"library_selection": "TODO",
"library_strategy": "TODO",
"fastq": [
{
"value": "single_cell_S1_L001_I1_001.fastq.gz",
"attributes": {
"read_type": "feature_barcode"
}
},
{
"value": "single_cell_S1_L001_R1_001.fastq.gz",
"attributes": {
"read_type": ["paired", "umi_barcode"]
}
},
{
"value": "single_cell_S1_L001_R2_001.fastq.gz",
"attributes": {
"read_type": "sample_barcode"
}
},
{
"value": "single_cell_S1_L001_R3_001.fastq.gz",
"attributes": {
"read_type": ["paired", "cell_barcode"]
}
}
]
}
Manifest File Types
Please refer to the more detailed documentation for supported file types for each submission.
Sequence based submission support the following formats:
FASTA: Sequences in fasta format
FLATFILE: Sequences in EMBL-Bank flat file format
The following additional formats are supported for genome assembly submissions:
AGP: Sequences in AGP format
CHROMOSOME_LIST: list of chromosomes
UNLOCALISED_LIST: list of unlocalised sequences
The following formats are supported for read submissions:
BAM: BAM file
CRAM: CRAM file
FASTQ: fastq file
Info File (for backward compability only)
You can also provide the metadata fields in a separate info file. The info file has the same format as the manifest file.
When a separate info file is used then the manifest file must contain the INFO
field pointing to the info file.
For example, the following manifest file represents a genome assembly consisting of contigs provided in one fasta file:
INFO assembly.info
FASTA genome.fasta.gz
Stage 3: Validate and Submit Files
You can validate your files using the -validate command line option. Note that
the -submit option must be used to submit the validated files.
You can submit your files using the -submit command line option. Before
being submitted your files will be validated and uploaded to your
private Webin file upload area in webin.ebi.ac.uk.
Please refer to individual modules for validation rules.
Validation error reports are written into the <outputDir>/<context>/<name>/validate directory.
The webin command line submission interface creates and submits XMLs for you. These XMLs and
the Receipt XML containing accession numbers are written into the <outputDir>/<context>/<name>/submit
directory. This directory also contains the file manifest that refers to the files that are
part of the submission.
The <outputDir> can be specified using the -outputDir option, the <context> is
specified using the -context option, and the <name> is a submitter provided unique
name specified in the manifest file.
Once the submission is complete an accession number is immediately returned to the submitter by the Webin command line submission interface. Please refer to individual modules for advice which long term stable accession numbers can be used in publications.
Output Directory Structure
An output directory can be specific to the Webin command line submission interface
using the -outputDir option. This directory will have the following subdirectories:
<context>/<name>/validate<context>/<name>/submit
If the -outputDir option is not specified then the directory in which the
-manifest file is used as the output directory.
The <context> is the submission type provided using the -context option
and the <name> is the unique name provided in the manifest file.
The
validatedirectory contains the validation reports created using the-validateoption.The
submitdirectory contains the XMLs created by the-submitoption including the Receipt XML. This directory also contains the file manifest that refers to the files that are part of the submission.
Validation Reports
If the -validate action fails for any reason then validation reports are written into directory:
<context>/<name>/validate
The validation reports correspond to the input files with an added suffix .report.
For example, a validated fasta file assembly.fasta will have a corresponding validation
report assembly.fasta.report.
Messages which can’t be attributed to a specific input file will be written to both standard out and in the following file:
<context>/<name>/validate/webin-cli.report
Run the program using the Docker image
Webin-CLI is available as the enasequence/webin-cli Docker image.
You can run the Webin-CLI docker image using docker:
docker run enasequence/webin-cli
or using singularity:
singularity run docker://enasequence/webin-cli
The required command line options are explained below. Please remember to mount local directories containing the files to submit so that they available to the running container.
Configuring Your Firewall For ENA Upload
Some users may encounter problems connecting to the ENA FTP service, a necessary step in Webin-CLI submission. A possible solution to this is to ensure that your firewall is configured appropriately to allow you to connect to this service:
Proxy Servers
If your organisation uses a https proxy you can set the following Java properties to instruct the webin-cli to use them:
https.proxyHost
https.proxyPort
For example:
java -Dhttps.proxyHost=proxy.com -Dhttps.proxyPort=8080 -jar webin-cli-<version>.jar <options>
Similarly, if your organisation uses a ftp proxy you can set the following properties:
ftp.proxyHost
ftp.proxyPort
For example:
java -Dftp.proxyHost=proxy.com -Dftp.proxyPort=8080 -jar webin-cli-<version>.jar <options>
Using Aspera Instead of FTP to Upload Files
By default the Webin command line interface will use FTP to upload files to the
webin.ebi.ac.uk server. Alternatively, you may use the Aspera protocol by installing
Aspera Cli and specifying the
-ascp option. Aspera is a commercial file transfer protocol that may provide better
transfer speeds than FTP making it useful when uploading larger files.
Please note that that the folder containing the ascp command line program
must be included in the PATH variable. The ascp command can be found from
the cli/bin directory of the downloaded and expanded Aspera Cli archive file.
Release policy
Webin-CLI uses standard three number semantic versioning.
Patch releases: the third digit is incremented by one after backward compatible bug fixes. For example, from 1.0.1 to 1.0.2.
Minor releases: the second digit is incremented by one after backward compatible new features. For example, from 1.0.1 to 1.1.0.
Major releases: changes that break backward compatibility result in the first digit being incremented. For example, from 1.0.1 to 2.0.0.
The definition of Webin-CLI backward compatibility is that there are no breaking changes to the command line usage or to the file formats.
All releases are made immediately after bugs have been fixed or new features have been added.
Releases are downloadable from: https://github.com/enasequence/webin-cli/releases.
After all releases, we will endeavour to contact affected submitters who previously were unable to complete their submissions.
Minor and Major releases will be announced to ena-announce@ebi.ac.uk mailing list.
After Minor or Major releases, submitters will be asked to migrate to use this (or higher) version after a transition period.
After a Minor release, we will give two weeks notice for submitters to migrate to the new (or higher) version.
After a Major release, we will give at least two months notice for submitters to migrate to the new (or higher) version.