Submitting Environmental Single-Cell Amplified Genomes
Introduction
Environmental SAG assemblies can be submitted to the European Nucleotide Archive (ENA) using the
Webin command line submission interface with -context genome.
Please contact our helpdesk if you intend to submit an assembly assembled from third party data.
Each SAG from an environmental source requires a derived SAG sample so please follow instructions carefully.
Genome assembly submissions include plasmids, organelles, complete virus genomes, viral segments/replicons, bacteriophages, prokaryotic and eukaryotic genomes.
An environmental SAG assembly consists of:
General assembly information
Study accession or unique name (alias)
Environmental Sample accession or unique name (alias)
SAG Sample accession or unique name (alias)
Assembly name
Assembly program
Sequencing platform
Minimum gap length
Molecule type (genomic DNA, genomic RNA or viral cRNA)
Coverage
Free text description of the assembly (optional)
Contig/Chromosome sequences
Functional annotation (optional)
For assembly submission purposes, the term ‘chromosome’ should be understood to include organelles (e.g. mitochondria and chloroplasts), plasmids and viral segments.
The below image illustrates the stages of the environmental SAG submission process:
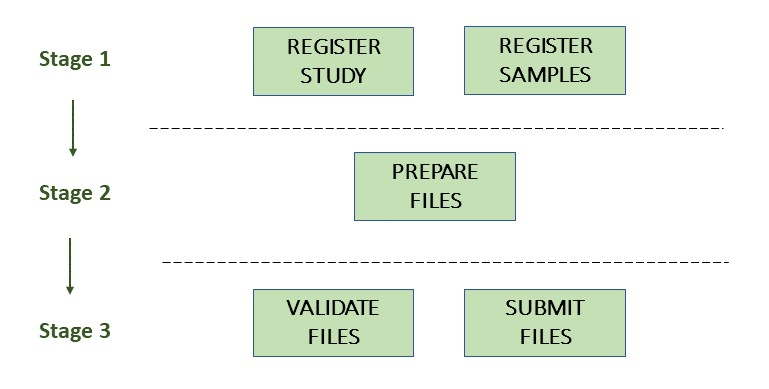
Stage 1: Pre-register study and samples
Each submission must be associated with a pre-registered study and a SAG sample.
Genome assemblies except primary metagenomes are uniquely associated with a study and a sample.
If you have not done so already, please register a study. If you intend to submit your SAG with annotation, make sure to register locus tag prefixes during this stage.
It is also strongly recommended to submit the reads from which the assembly was assembled. When submitting the raw reads in a Single-Cell Amplification study, these should either be in multi-fastq format or remain multiplexed in BAM format, and be submitted to an environmental sample. This sample should use the most appropriate environmental checklist and an environmental taxon (e.g. aquatic metagenome (tax id: 1169740)).
If you do not intend to submit raw reads as part of your SAG submission, please make sure you have registered environmental samples anyway. These will represents the original sequenced biomaterial that your SAG was derived from. You will also need to follow these guidelines for details on how to release your environmental samples. If no data is associated with a sample, it needs to be released manually in order to be available to the public.
Registering SAG samples
Each SAG assembly submission must be associated with a SAG sample. This is because a SAG is not an assembly of all the data from the collected sample so linking to the environmental sample is misleading and causes incorrect taxonomy assignment. These SAG samples represent the individual organisms derived from the environmental sample and hold all metadata related to the taxonomy of that subset as well as methods used to derive it.
SAG samples should be as specific in taxonomy as they can be and use the specific GSC MISAGS checklist.
Please make sure these SAG samples correctly reference the environmental sample that the SAG was derived from. This can be done from within the checklist using the mandatory “sample derived from” attribute. If the assembly was derived from multiple samples or runs you can list these with a comma separated list or range.
You should also reference the source sample in the description:
“This sample represents a Single-Cell Amplified Genome derived from the environmental sample ERSXXXXX”
Stage 2: Prepare the files
The set of files that are part of the submission are specified using a manifest file.
The manifest file is specified using the -manifest <filename> option.
The files required for submission of a genome assembly depends on the assembly level. Compared to more complex genomes which can be submitted as contigs, scaffolds or chromosomes, SAGs are usually only submitted at one of two levels:
Contig assembly
Consists of the following files:
1 manifest file
1 FASTA file OR 1 flat file
This assembly level only requires information on the sequences and annotation (if any).
You will receive an error if less than 2 or more than 1,000,000 sequences are submitted. If your assembly is a single sequence and the genome is highly complete, please submit this as a Chromosome assembly. If the sequence is not complete enough to consider it a fully assembled chromosome, you can request your assembly be considered as a ‘single contig’ assembly through our helpdesk.
Chromosome assembly
Consists of the following files:
1 manifest file
1 FASTA file OR 1 flat file
This assembly level allows the submission of fully assembled chromosomes (including organelles, plasmids, and viral segments). This requires information on the sequences and annotation (if any) and submission of a chromosome list file to indicate which sequences represent which ‘chromosomes’.
If these chromosomes contain unlocalised sequences (where the chromosome of the sequence is known but not the exact location) you can submit an additional unlocalised list file.
Sequence names
Sequences must have a unique name within the submission that is provided in the fasta or flat files. It is essential that the sequence names are unique and used consistently between files.
For example, the chromosome list file must refer to the chromosome sequences using the unique sequence names.
Manifest file
The manifest file has two columns separated by a tab (or any whitespace characters): - Field name (first column): case insensitive field name - Field value (second column): field value
The following metadata fields are supported in the manifest file:
STUDY: Study accession or unique name (alias)
SAMPLE: SAG Sample accession or unique name (alias)
ASSEMBLYNAME: Unique assembly name
ASSEMBLY_TYPE: ‘Environmental Single-Cell Amplified Genome (SAG)’
COVERAGE: The estimated depth of sequencing coverage
PROGRAM: The assembly program
PLATFORM: The sequencing platform, or comma-separated list of platforms
MINGAPLENGTH: Minimum length of consecutive Ns to be considered a gap (optional)
MOLECULETYPE: ‘genomic DNA’, ‘genomic RNA’ or ‘viral cRNA’ (optional)
DESCRIPTION: Free text description of the genome assembly (optional)
RUN_REF: Comma separated list of run accession(s) (optional)
Please see further below for validation rules affecting some of these fields.
The following file name fields are supported in the manifest file:
FASTA: sequences in fasta format
FLATFILE: sequences in EMBL-Bank flat file format
AGP: Sequences in AGP format
CHROMOSOME_LIST: list of chromosomes
For example, the following manifest file represents an environmental single-cell assembly consisting of contigs provided in one FASTA file:
STUDY TODO
SAMPLE TODO
RUN_REF TODO
ASSEMBLYNAME TODO
ASSEMBLY_TYPE Environmental Single-Cell Amplified Genome (SAG)
COVERAGE TODO
PROGRAM TODO
PLATFORM TODO
MINGAPLENGTH TODO
MOLECULETYPE genomic DNA
FASTA single-cell_genome.fasta.gz
Stage 3: Validate and submit the files
Files are validated, uploaded and submitted using the Webin command line submission interface (Webin-CLI). Please refer to the Webin command line submission interface documentation for full information about the submission process.
Brief examples of Webin-CLI commands follow.
The tool has -submit and -validate options which are mutually exclusive.
Full validation of your data and metadata is run regardless of which option you choose, but using just -validate
gives you the opportunity to check the validation of your assembly and information on any errors.
You are therefore encouraged to make use of Webin-CLI validation as much as you need to before you are ready to submit
for real.
First, run the Webin-CLI validation command, specifying your credentials and the path to your manifest file:
webin-cli -username Webin-XXXXX -password YYYYYYY -context genome -manifest manifest.txt -validate
Second, run the Webin-CLI submission command:
webin-cli -username Webin-XXXXX -password YYYYYYY -context genome -manifest manifest.txt -validate
In both cases, your prospective submission will be validated in full, and the result of this reported to you. A successful validation results in a simple success message, while a successful submission will further result in the assigned accession number (see below) being reported at your command line. Meanwhile, a failed validation will provide direction to a report file where you can find a list of error messages explaining the reason for the failure, which you can address before re-attempting.
For more information on how to install and use Webin-CLI, please refer to the Webin-CLI Submission page.
Assigned accession numbers
Once the genome assembly has been submitted an analysis (ERZxxxxxx) accession number is immediately assigned and returned to the submitter by the Webin command line submission interface (Webin-CLI).
ERZ accessions should not be used to reference the assembly in publications. The purpose of the ERZ accession number is for the submitter to be able to refer to their submission within the Webin submission service. For example, the submitter can retrieve the assigned genome assembly and sequence accessions from the Webin Portal or from the Webin reports service using the ERZ accession number. This accession should be used to refer to the assembly in any conversations with helpdesk staff.
For Environmental Single-cell Amplified Genome assemblies, long term stable accession numbers that can be used in publications are:
Study accession (PRJEBxxxxx) assigned at time of study registration.
Sample accession (SAMEAxxxxxx) assigned at time of sample registration.
Genome assembly accession (GCA_xxxxxxx) assigned once the assembly has been fully processed by ENA.
Sequence accession(s) assigned once the assembly has been fully processed by ENA.
Submitters can retrieve the genome and sequence accession numbers from the Webin Portal or from the Webin reports service. These accession numbers are also sent to the submitters by e-mail.
Validation rules
Assembly submissions are subject to a great deal of validation before submission is allowed. Some key points are described here.
Sample And Study Validation
Sample and study (BioProject) pair must be unique for an assembly
Sample taxonomic classification must be species rank or below (or equivalent) within NCBI taxonomy.
Assembly name validation
Assembly names must:
match the pattern: ^[A-Za-z0-9][A-Za-z0-9 _#-.]*$
not be longer than 50 characters
not include the name of the organism assembled
Chromosome name validation
Chromosome names must:
match the pattern: ^[A-Za-z0-9][A-Za-z0-9_#-.]*$
be shorter than 33 characters
- not contain any of the following as part of their name (case insensitive):
‘chr’
‘chrm’
‘chrom’
‘chromosome’
‘linkage group’
‘linkage-group’
‘linkage_group’
‘plasmid’
be unique within an assembly
Sequence validation
Sequences must: - have unique names within an assembly - be at least 20bp long - not have terminal Ns - consist of bases: ‘a’,’c’,’g’,’t’,’u’,’b’,’d’,’h’,’k’,’m’,’n’,’r’,’s’,’v’,’w’,’y’