Submitting A Primary Metagenome Assembly
Introduction
Metagenome assemblies can be submitted to the European Nucleotide Archive (ENA) using
the Webin command line submission interface with -context genome.
Please contact our helpdesk if you intend to submit an assembly assembled from third party data.
A primary metagenome assembly is a metagenome assembly prior to binning from a sampled biome or collection of sampled biomes without attempt to separate taxa.
A primary metagenome assembly consists of:
General assembly information
Study accession or unique name (alias)
Environmental Sample accession or unique name (alias)
Assembly program
Sequencing platform
Minimum gap length
Molecule type (genomic DNA, genomic RNA or viral cRNA)
Coverage
Free text description of the assembly (optional)
Contig sequences in fasta format
The following picture illustrates the stages of the metagenome assembly submission process:
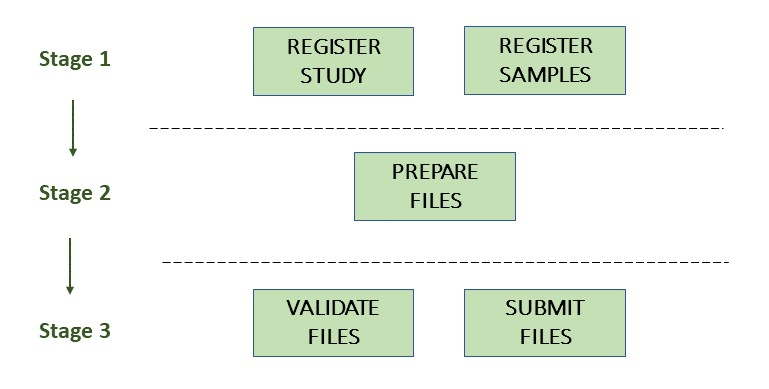
Stage 1: Pre-register study and environmental sample
Each submission must be associated with a pre-registered study and environmental sample. This sample should be the same sample used to submit your raw reads.
When registering an environmental sample, please make sure the appropriate environmental checklist is chosen for this and an environmental taxon is used (e.g. aquatic metagenome (tax id: 1169740)).
The methods for submitting metagenomic studies and samples follow the same process as any other study/sample submission. Follow the links for more information.
It is strongly recommended to submit and reference raw reads associated with the assembly being submitted.
In order to reference the reads which were used to generate the assembly, please see the RUN_REF tag included in
the manifest file below.
Stage 2: Prepare the files
The set of files that are part of the submission are specified using a manifest file.
The manifest file is specified using the -manifest <filename> option.
A primary metagenome assembly submission consists of the following files:
1 manifest file
1 fasta file
Manifest file
The manifest file has two columns separated by a tab (or any whitespace characters):
Field name (first column): case insensitive field name
Field value (second column): field value
The following metadata fields are supported in the manifest file:
STUDY: Study accession or unique name (alias)
SAMPLE: Environmental sample accession or unique name (alias)
ASSEMBLYNAME: Unique assembly name
ASSEMBLY_TYPE: ‘primary metagenome’
COVERAGE: The estimated depth of sequencing coverage
PROGRAM: The assembly program
PLATFORM: The sequencing platform, or comma-separated list of platforms
MINGAPLENGTH: Minimum length of consecutive Ns to be considered a gap (optional)
MOLECULETYPE: ‘genomic DNA’, ‘genomic RNA’ or ‘viral cRNA’ (optional)
DESCRIPTION: Free text description of the genome assembly (optional)
RUN_REF: Comma separated list of run accession(s) (optional)
Please see further below for validation rules affecting some of these fields.
The following file name fields are supported in the manifest file for a primary metagenome submission:
FASTA: sequences in fasta format
For example, the following manifest file represents a primary metagenome assembly consisting of contigs provided in one fasta file:
STUDY TODO
SAMPLE TODO
RUN_REF TODO
ASSEMBLYNAME TODO
ASSEMBLY_TYPE primary metagenome
COVERAGE TODO
PROGRAM TODO
PLATFORM TODO
MINGAPLENGTH TODO
MOLECULETYPE genomic DNA
FASTA primary_metagenome.fasta.gz
Stage 3: Validate and submit the files
Files are validated, uploaded and submitted using the Webin command line submission interface (Webin-CLI). Please refer to the Webin command line submission interface documentation for full information about the submission process.
Brief examples of Webin-CLI commands follow.
The tool has -submit and -validate options which are mutually exclusive.
Full validation of your data and metadata is run regardless of which option you choose, but using just -validate
gives you the opportunity to check the validation of your assembly and information on any errors.
You are therefore encouraged to make use of Webin-CLI validation as much as you need to before you are ready to submit
for real.
First, run the Webin-CLI validation command, specifying your credentials and the path to your manifest file:
webin-cli -username Webin-XXXXX -password YYYYYYY -context genome -manifest manifest.txt -validate
Second, run the Webin-CLI submission command:
webin-cli -username Webin-XXXXX -password YYYYYYY -context genome -manifest manifest.txt -validate
In both cases, your prospective submission will be validated in full, and the result of this reported to you. A successful validation results in a simple success message, while a successful submission will further result in the assigned accession number (see below) being reported at your command line. Meanwhile, a failed validation will provide direction to a report file where you can find a list of error messages explaining the reason for the failure, which you can address before re-attempting.
For more information on how to install and use Webin-CLI, please refer to the Webin-CLI Submission page.
Assigned accession numbers
Once the genome assembly has been submitted an analysis (ERZxxxxxx) accession number is immediately assigned and returned to the submitter by the Webin command line submission interface.
The purpose of the ERZ accession number is for the submitter to be able to refer to their submission within the Webin submission service and access their data in the browser.
For primary metagenome assemblies, long term stable accession numbers that can be used in publications are:
Study accession (PRJEBxxxxx) assigned at time of study registration.
Sample accession (SAMEAxxxxxx) assigned at time of sample registration.
See an example of a publicly available primary metagenome at: https://www.ebi.ac.uk/ena/browser/view/ERZ1091679
Validation rules
Assembly name validation
Assembly names must:
match the pattern: ^[A-Za-z0-9][A-Za-z0-9 _#-.]*$
not be longer than 50 characters
not include the taxonomic name of the organism assembled
Sequence validation
Sequences must:
have unique names within an assembly
be at least 20bp long
not have terminal Ns
consist of bases: ‘a’,’c’,’g’,’t’,’u’,’b’,’d’,’h’,’k’,’m’,’n’,’r’,’s’,’v’,’w’,’y’